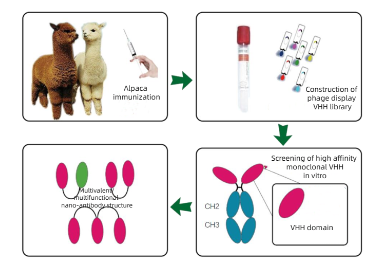
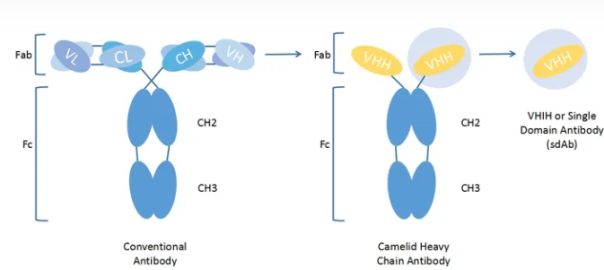
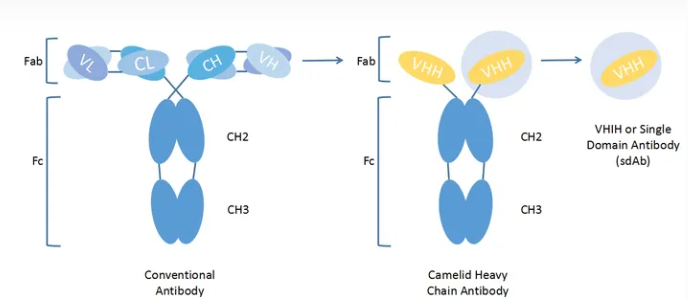
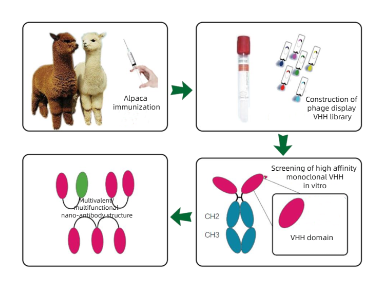
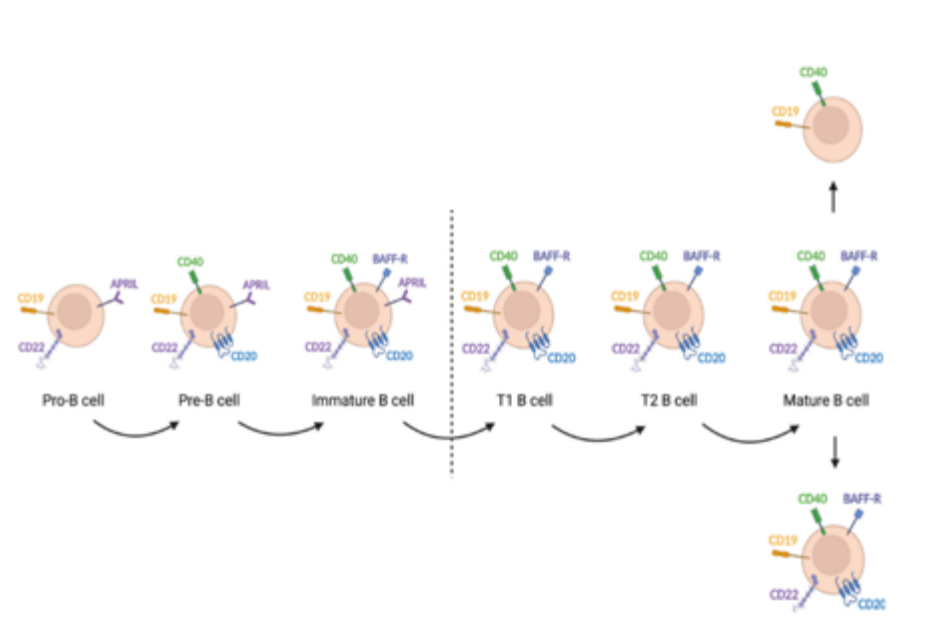
Alpaca Immune isolation What cells are isolated by PBMC?
The isolated peripheral blood mononuclear cells are the main cell types of mononuclear cells in the blood, mainly including lymphocytes (T /B), monocytes, phagocytes, dendritic cells, and other small numbers of cell types, of which lymphocytes account for a large part.
What is the purpose of PBMC separation?
The main purpose of isolating PBMC is to remove multinucleated cells and red blood cells so that it can easily simulate the blood immune environment in vitro. Preparation of PBMC from blood is a common step before isolating a specific subpopulation of immune cells.
What are the precautions during PBMC separation?
(1) Not applicable to abnormal samples such as blood clotting or samples over 48 hours.
(2) It is recommended to use 4-9ml samples for 15ml separation tubes; For 50ml separation tubes, 13-30ml samples are recommended.
(3) For samples placed for more than 24 hours, it is recommended to lengthen the centrifugation time.
(4) Before harvesting cells after centrifugation, the upper plasma layer can also be absorbed, collected, or discarded to help prevent platelet contamination.
(5) When pouring supernatant after centrifugation, do not invert the separation tube for more than 2s.
(6) Do not reuse the separation pipe.
(7) After centrifugation, cells may gather on the wall of the separation tube above the enrichment layer. This polymerization is normal and is affected by sample quality, sample placement time, and type of anticoagulant. This polymerization is independent of the use of the separation tube. The cells can be removed by lightly scraping one side of the cluster using the pipette tip.
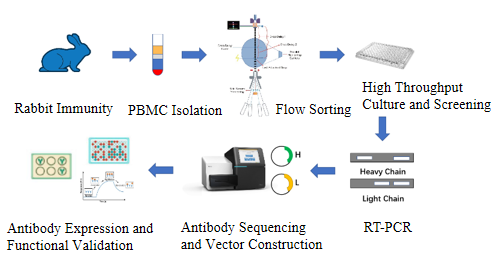
How can alpaca Immunology obtain highly effective serum and high-affinity antibodies through follow-up screening?
High-quality immunogen preparation
the purity of the immunogen should be as high as possible to avoid non-specific reactions. For the protein immunogen, it is ensured that it remains intact in three-dimensional structure to better simulate the antigen epitope in the natural case. If the target is a surface antigen or receptor, consider using a recombinant protein that is full-length, complex, or expressed in an appropriate expression system.
Selection and use of adjuvants
Use adjuvants that can enhance long-term immune memory and stimulate the production of high antibody titers, such as Freund’s complete or incomplete adjuvants, hydrated aluminum hydroxide, etc. In some cases, to further enhance the immune response, specific immune-stimulating complexes (such as CpG oligonucleotides) may be added to the adjuvant.
Design of immunization program
Establish a reasonable immunization schedule, including basic immunization, multiple booster immunization, and give enough time after each immunization to allow the immune system to fully respond. Blood is taken at appropriate times and antibody levels are monitored to determine whether additional booster immunization or adjusted immunization regimen is required.
Serum screening and affinity maturation of antibodies
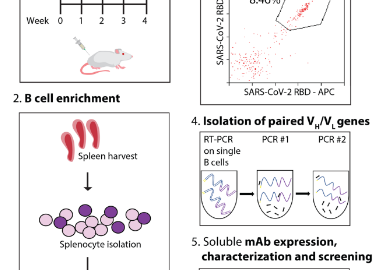
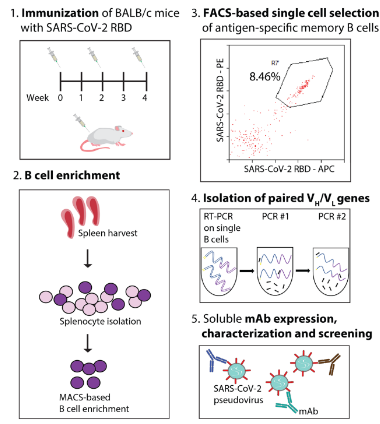
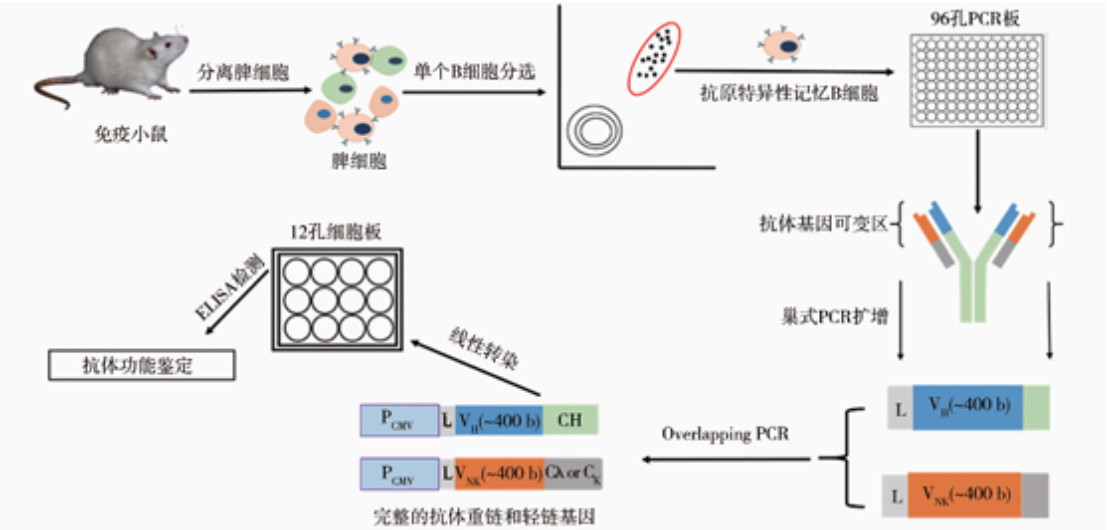
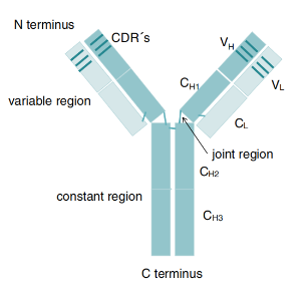
ELISA, Western blotting or immunoprecipitation are used to screen high-valence serum samples. For situations where monoclonal antibodies need to be generated, B cells can be isolated from immunized alpacas, single-cell screening can be performed, and then specific antibody genes can be cloned using molecular biological methods (such as PCR, plasmid construction, etc.). If needed, the affinity of the resulting antibody can be enhanced by in vitro affinity maturation techniques (e.g., phage-based or yeast-based display techniques).
Production and validation of high-affinity antibodies
Use of suitable expression systems (e.g. mammalian cells, E. coli, etc.) to produce recombinant antibodies. Antibodies are purified and thoroughly analyzed for biological function and binding properties to ensure that they have the expected high affinity and specificity.
References
[1] Cortez-Retamozo V .Efficient Cancer Therapy with a Nanobody-Based Conjugate[J].Cancer Research, 2004, 64(8):2853-2857.DOI:10.1158/0008-5472.CAN-03-3935.
[2] Meyer T D , Muyldermans S , Depicker A .Nanobody-based products as research and diagnostic tools[J].Trends in Biotechnology, 2014, 32(5).DOI:10.1016/j.tibtech.2014.03.001.
[3] Steyaert J , Kobilka B K .Nanobody stabilization of G protein-coupled receptor conformational states.[J].Current Opinion in Structural Biology, 2011, 21(4):567-572.DOI:10.1016/j.sbi.2011.06.011.