Bacteriophages, or simply phages, are viruses that specifically infect and replicate within bacteria. Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome and may have structures that are either simple or elaborate. They were discovered independently by Frederick Twort in 1915 and Félix d’Hérelle in 1917 [1]. Phages are among the most abundant and diverse entities in the biosphere, with an estimated 10^31 phages on Earth. They play critical roles in microbial ecology, evolution, and even in human health by influencing bacterial populations.
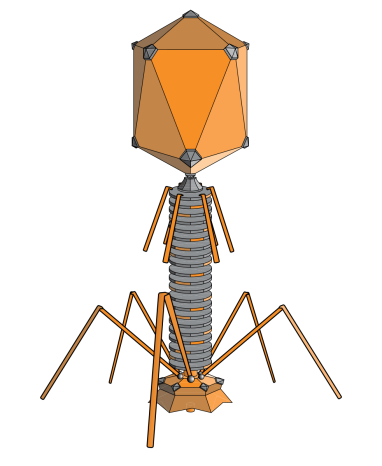
(Figure 1: Common Phage Structure. Figure Source: Wikipedia)
Commonly studied bacteriophages
The classification of phages is complex and numerous. Among countless phages, only a few have been studied in detail. Let’s list a few types of phages to introduce them:
λ phage
Enterobacteria phage λ (lambda phage, coliphage λ, officially Escherichia virus Lambda) is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli (E. coli).λ Phage is a well-studied temperate bacteriophage that infects the bacterium Escherichia coli (E. coli). Discovered by Esther Lederberg in 1950, it is a key model organism in molecular genetics [2].λ phage has a double-stranded DNA (dsDNA) genome of approximately 48.5 kilobases.λ phage is widely used in molecular cloning and gene regulation studies due to its well-characterized genetics and the ability to integrate into the host genome.
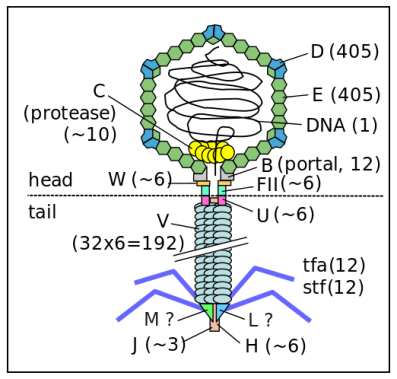
(Figure 2: Structure of λ Phage. Figure Source: Wikipedia)
M13 phage
M13 Phage is a filamentous bacteriophage that specifically infects male (F+) strains of E. coli by binding to the F pilus. M13 has a single-stranded DNA (ssDNA) genome of about 6.4 kilobases [3]. M13 is commonly used in phage display technology for protein engineering and antibody development. Its ability to display peptides and proteins on its surface makes it a valuable tool in biotechnological applications.
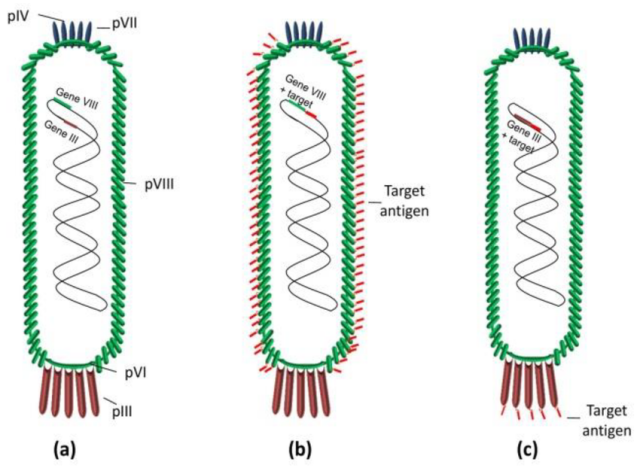
( Figure 3:Scheme of bacteriophage M13: (a) wild-type phage; (b) phage displaying multivalent copies of the target antigen on pVIII protein; and (c) phage displaying multivalent copies of the target antigen on phage pIII protein. In both cases, the DNA fragments encoding the target antigens were cloned into a phage vector [4].)
T4 phage
T4 Phage is a lytic bacteriophage that infects E. coli. It is one of the largest and most complex phages studied. T4 has a large dsDNA genome of approximately 169 kilobases. T4 is a relatively large virus, approximately 90 nm wide and 200 nm long (most viruses range from 25 to 200 nm in length) [5]. T4 phage is used as a model system for studying DNA replication, recombination, and repair. Its well-defined life cycle and genetic system make it ideal for these studies.
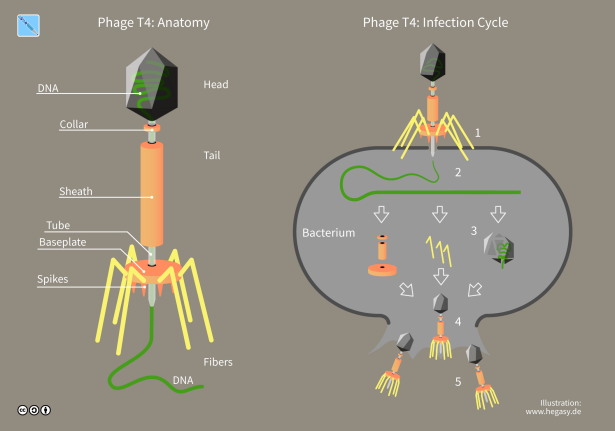
(Figure 4: Structure of T4 phage. Figure Source: Wikipedia)
T7 phage
Bacteriophage T7 (or the T7 phage) is a bacteriophage, a virus that infects bacteria. It infects most strains of Escherichia coli and relies on these hosts to propagate. It is known for its simplicity and rapid replication cycle. T7 has a linear dsDNA genome of about 40 kilobases. It is extensively used in molecular biology, particularly in the T7 expression system for high-level protein expression. The T7 RNA polymerase is highly specific and efficient, making it a powerful tool for gene expression studies.
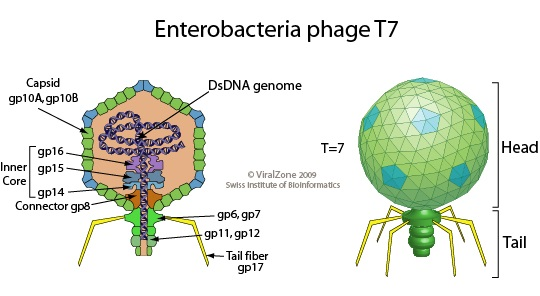
(Figure 5: Structure of T7 phage. Figure Source: Wikipedia)
Phage display technology is an in vitro antibody screening method developed by George P. Smith in 1985. The principle is to insert genes encoding exogenous peptides or proteins into the appropriate positions of the structure genes of the bacteriophage’s outer shell protein. With normal reading frames and without affecting the normal function of the outer shell protein, the exogenous peptides or proteins form fusion proteins on the bacteriophage’s outer shell protein, which is presented on the surface of the bacteriophage as the offspring bacteriophage reassembles. The displayed protein can maintain a relatively independent spatial structure and biological activity, which is conducive to the binding of target proteins. Therefore, target proteins can be quickly used for multi-round screening of phage display antibody libraries and expanded cultivation in E. coli, a process also known as “panning”. Repeated screening can gradually increase the proportion of phages that specifically recognize target molecules, ultimately obtaining peptides or proteins that recognize target molecules.
Reference
Ackermann, H. W. (2009). “Phage Classification and Characterization.” Methods in Molecular Biology, 501, 127-140.
Ptashne, M. (2004). “A Genetic Switch: Phage Lambda Revisited.” Cold Spring Harbor Laboratory Press.
Smith, G. P., & Petrenko, V. A. (1997). “Phage Display.” Chemical Reviews, 97(2), 391-410.
Palma, Marco. (2023). Aspects of Phage-Based Vaccines for Protein and Epitope Immunization. Vaccines. 11. 436. 10.3390/vaccines11020436.
Miller, E. S., et al. (2003). “Bacteriophage T4 Genome.” Microbiology and Molecular Biology Reviews, 67(1), 86-156.
